Research Highlights
Research highlights articles and press releases between November 2019 to February 2020
Single motor proteins visualized in action for the first time
Feb. 07, 2020
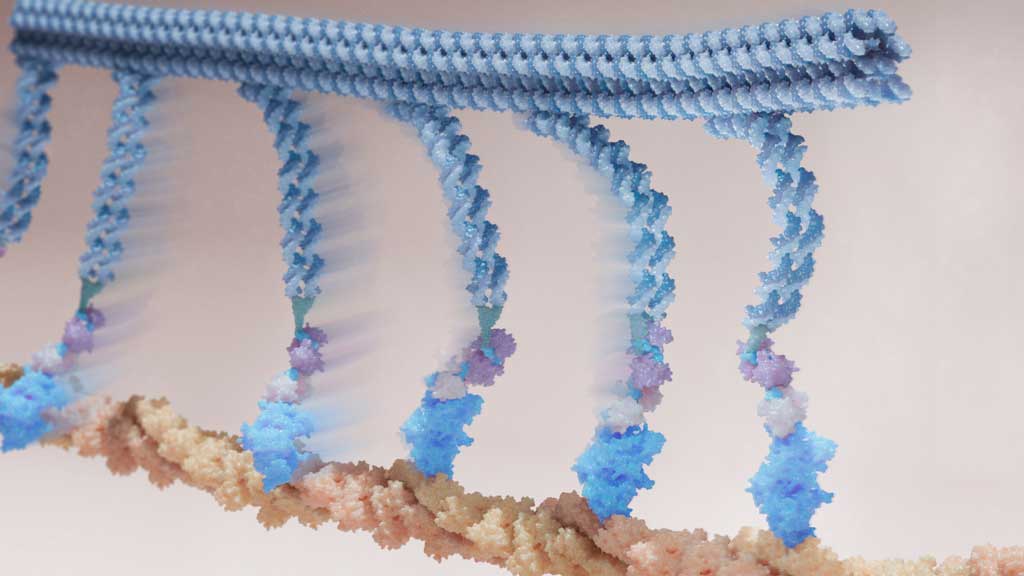
Muscles contract by sliding between two types of filaments—thick filaments of myosin and thin filaments of actin. But there has been a long-standing controversy about the dynamics of this molecular machine. A team led by Mitsuhiro Iwaki of Lab for Cell Dynamics Research have now directly visualized the individual myosin molecules in action. Using DNA origami technology, they first created rod structures resembling thick filaments of muscle tissue to which they attached myosin proteins along this rod in precise positions (top image). Then, using advanced single-molecule imaging, they snapped atomic-scale pictures of myosin interacting with actin under biologically realistic geometric conditions. Their achievement could aid in the hunt for new ways to treat diseases associated with myosin malfunction. More
Fujita K, Ohmachi M, Ikezaki K, et al. Commun Biol 2, 437 (2019)
Faster modeling of interactions between ligands and proteins
Nov. 29, 2019
Many drugs work by mimicking the natural interactions of small molecules known as ligands and large proteins involved in biological processes, thus, it is vital to accurately simulate such interactions when designing new drugs. Now, Yuji Sugita and Suyong Re in the Laboratory for Biomolecular Function Simulation and others have extended an existing molecular dynamics simulation method, allowing them to compute hundreds of binding and unbinding events between a protein and a ligand in about a month using the K computer, whereas conventional methods would take at least ten times longer. Their improved method promises to boost the speed and accuracy of designing new drugs. More
Re S, Oshima H, Kasahara K, et al. Proc Natl Acad Sci USA 116, 18404 (2019)

New supercomputer dedicated to drug discovery
Nov. 29, 2019
A RIKEN-wide collaborative team spearheaded by Team Leader Makoto Taiji of the Laboratory for Computational Molecular Design developed a new supercomputer, called MDGRAPE-4A, which is designed specifically for molecular dynamics simulations. MDGRAPE-4A, located at the BDR Osaka Campus, has an overall power of 1.3 petaflops and can outperform speedier general-purpose supercomputers due to its dedicated architecture. The supercomputer can model the interactions between drugs and proteins in the body, and understanding these interactions will make it possible to develop more effective molecular targeted therapies. More
Detecting the metabolites of a single cell
Nov. 15, 2019
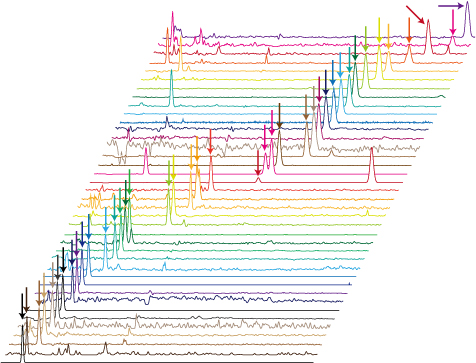
Cells are constantly manufacturing the compounds they need to survive and thrive. While scientists can determine the genetic makeup of and the messenger RNA produced by a single cell, there was no reliable technique for profiling the small molecules, or metabolites, produced by a single cell. Such a technique would provide valuable information for cancer biology, as cancer cells acquire different strategies for survival, thus, showing a different metabolite profile. A team led by Takayuki Kawai (Laboratory for Single Cell Mass Spectrometry) has now boosted the sensitivity of a conventional metabolomics analytical technique called capillary electrophoresis-mass spectrometry by up to 800 times, allowing them identify 40 metabolites from a single human cervical cancer cell. The team is now searching for good target molecules for clinical applications, such as drug discovery and diagnosis. More
Kawai T, Ota N, Okada K, et al. Anal Chem 91, 10564 (2019)