2020-08-27
Research Highlights
Research highlights articles and press releases between March 2020 to July 2020

Mice need kinetochores rich in a microtubule crosslinker to achieve error-free oocyte division
Jul. 31, 2020
Animal oocytes are susceptible to errors during the segregation of genetic material. These errors can result in miscarriages and congenital disorders such as Down syndrome. Tomoya Kitajima and Shuhei Yoshida from the Lab for Chromosome Segregation and collaborators have found that mice need kinetochores (the main point of attachment for spindle microtubules) rich in the microtubule crosslinker Prc1 to achieve error-free formation of spindles during oocyte cell division. Significantly, the team found that kinetochores in humans are not rich in Prc1. This difference between mouse and human oocytes could go a long way to explaining why oocyte division in humans is more error prone than that in mice. More
Yoshida S, Nishiyama S, Lister L, et al. Nat Commun 11, 2652 (2020)
A new biomarker for the aging brain
May 18, 2020
Toshihiko Aso, deputy team leader of Lab for Brain Connectomics Imaging and his collaborators have identified changes in the aging brain related to blood circulation. In their study, they found that natural age-related enlargement of the ventricles, a condition called ventriculomegaly, was associated with a lag in blood drainage from a specific deep region of the brain (top image). The lag can be detected easily with magnetic resonance imaging (MRI), making it a potential biomarker for predicting ventriculomegaly and the aging brain, which can then be treated quickly. More
Aso T, Sugihara G, Murai T, et al. Brain 143, 1843-1856 (2020)
Finding more chromosome structures by assuming less
May 1, 2020
Some talk to people in their neighborhood, while others talk to people outside their communities but still in the same city. Likewise, chromosomes possess different types of structural features–closely interacting structures on a small scale are called TADs and longer range interacting structures on a larger scale are called compartments. Now, Yuichi Taniguchi, Vipin Kumar and Simon Leclerc in the Lab for Cell Systems Control have developed a new clustering technique for analyzing data derived from Hi-C experiments that allow them to identify multiple structural features at different scales at once and to associate them with a hierarchical tree. More
Kumar V, Leclerc S, Taniguchi Y. Nucleic Acids Res 48, e26 (2020)
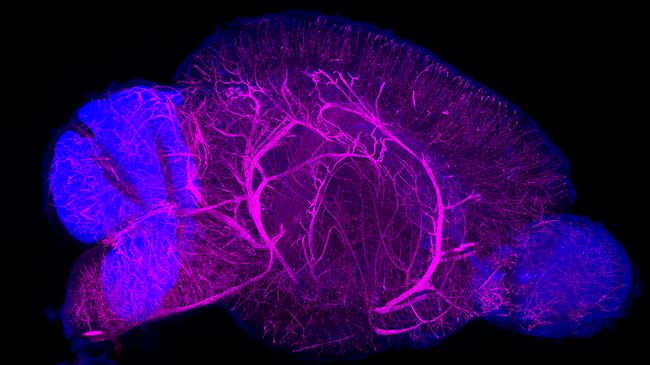
New staining technique visualizes whole organs and bodies
Apr. 27, 2020
Hiroki Ueda, Etsuo Susaki and colleagues from the Lab for Synthetic Biology have established an optimized three-dimensional (3D) tissue-staining and observation technique, named CUBIC-HistoVIsion, based on existing tissue clearing technology the lab previously developed. Their recent study details how the new technique can be used to stain tissue and label cells in mouse brains, human brains, and whole marmoset bodies. This technique will allow detailed anatomical analysis and whole-organ comparisons between species at the cellular level. More
Susaki EA, Shimizu C, Kuno A, et al. Nat Commun 11, 1982 (2020)
RIKEN group leads world in single-cell transcriptome profiling
Apr. 7, 2020
Single-cell RNA sequencing (scRNA-seq), a technique used to characterize the transcriptome of individual cells in a sample, is currently a focus of intense research. Many scRNA-seq protocols are now available, but as they all have marked differences in standards an international group has benchmarked 13 different methods using a unified reference sample resource. The group, led by Holger Heyn of the Centro Nacional de Análisis Genómico in Spain, found that the Quartz-seq2 method, developed by Itoshi Nikaido‘s Lab for Bioinformatics Research, was overall the best method developed to sequence single-cell RNA. More
Mereu E, Lafzi A, Moutinho C, et al. Nat Biotechnol 38, 747-755 (2020)
Exploring the molecular dynamics of the new coronavirus
Mar. 23, 2020
Makoto Taiji and his colleagues in the Lab for Computational Molecular Design used the MDGRAPE-4A dedicated drug discovery supercomputer to analyze the structural dynamics of the main protease of SARS-CoV-2, the virus that causes the new coronavirus disease, COVID-19. The raw data of the 10 microsecond-long simulation has been published on Mendeley Data (doi:10.17632/vpps4vhryg.2) for use by researchers around the world. More
Stem cells exert tight control over the timing of brain development
Mar. 13, 2020
Brain stem cells, or radial glia, give rise to all the neurons in the cerebral cortex, and are stretched between the apical and basal membrane surfaces. The transition of some radial glia from symmetrical to asymmetrical division that later leads to brain expansion has been thought to be governed by changes in the mitotic spindle orientation in radial glia relative to the apical surface. But a new study by Fumio Matsuzaki and Ikumi Fujita in the Lab for Cell Asymmetry showed that while manipulating spindle orientation of early-stage radial glia did cause the radial glia to detach from the apical surface, it did not necessarily lead to their migration. Instead, early-stage radial glia can extend ‘endfeet’ that reattach to the apical surface allowing symmetric division to proceed. This discovery could also shed new insights into our understanding of mammalian brain evolution. More
Fujita I, Shitamukai A, Kusumoto F, et al. Nat Cell Biol 22, 26-37 (2020)
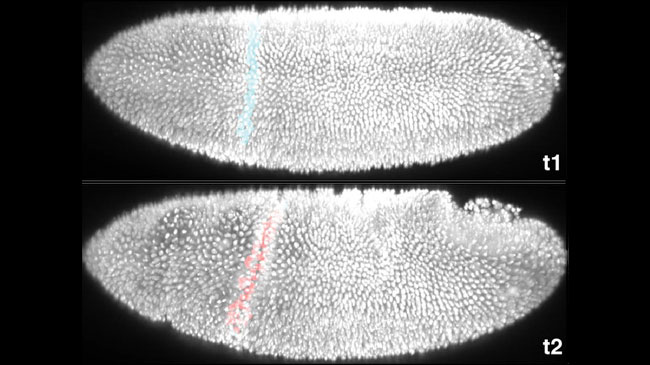
Mechanical forces shape animal “origami” precisely despite “noise”
Mar. 13, 2020
The reproducibility of form, shape, and characteristic appearance is a key feature of our development that is made possible because their instructions are coded in our DNA. What is perplexing, however, is how this reproducibility is achieved despite genetic variation and developmental “noise” resulting from environmental, physical and chemical fluctuations. An international team led by Yu-Chiun Wang (Lab for Epithelial Morphogenesis) discovered that a genetic “blueprint” for tissue bending, despite specific instructions down to the single-cell level, is insufficient to explain developmental consistency. The mechanical forces that sculpt the embryo turn out to be the noise-producing culprit, and unexpectedly, play a previously overlooked role that ensures the precision in tissue bending – a true double-edged sword! More
Eritano AS, Bromley CL, Bolea Albero A, et al. Dev Cell 53, 212-228.e12 (2020)