Approaching the Origin of Life with Structural Biology
It has been a while since I last talked to a structural biology researcher. I know that they use instruments like nuclear magnetic resonance (NMR) spectrometers to determine the structure of proteins, but I was very excited to be able to hear the story of what happens after the structure is determined as it sounded quite interesting.
Protein evolution?
You are part of Dr. Shunsuke Tagami’s lab, right? Which means that your research is related to the structural analysis of DNA and proteins?
I am doing structural biology, but I’m more focused on the origin of life.
My target is proteins. Proteins are the elements that are the driving force of all kinds of biological phenomena, and I am trying to uncover how modern proteins emerged and evolved.
Proteins evolved…?
Modern proteins are made up of about 300 or 400 amino acid residues that are strung together like beads. Some of the larger proteins are made up of over 1,000 amino acid residues. But ancient proteins were likely much smaller. The smallest proteins today are about one hundred amino acids in length. But even if they are only one hundred residues long, there are 20 types of amino acids, so the number of possible variations is 20100, or roughly 10130.
That’s an astronomical number.
Incidentally, the number of all atoms in the universe is considered to be 10 to the power of 80.
More than the number of atoms in the universe…
Yes, there are so many possible variations. However, in some proteins, only a few mutations can render them useless. This means that while there are a lot of variations, there are also many useless sequences. It’s almost a miracle that living organisms have been able to select only useful proteins from 10130 different variations.
Protein information is encoded in a DNA sequence, which is then transcribed into RNA, and RNA is further translated into a protein. So, I think that DNA and proteins need to evolve at the same time. It’s a bit like a chicken and egg situation, isn’t it?
That’s right. One of the important molecules in this chicken and egg situation is RNA polymerase, which is responsible for transcription. My research theme is to unravel the evolution of this enzyme.
What about the molecule that translates the RNA sequences into proteins?
The main player in translation is the ribosome. The ribosome is basically made of RNA with some proteins. When pondering about the origin of life, we have to consider the co-evolution of ribosomes and RNA polymerases. By the way, have you ever heard of the “RNA world” hypothesis?
I have. It is one hypothesis for the origin of life which proposes that RNA was used to carry out a range of different functions, right?
Yes, that’s right. So, there are a lot of people working on ribosomes, which are mainly composed of RNA.
But you are working on the RNA polymerase.
Targeting the core of RNA polymerase
We are trying to uncover the primitive form of RNA polymerase. RNA polymerase today is huge, but its catalytic core is composed of a small protein domain called double-psi beta-barrel (DPBB), which is about 80 amino acids in length. This DPBB structure is not only found in RNA polymerase, but in many other proteins as well.
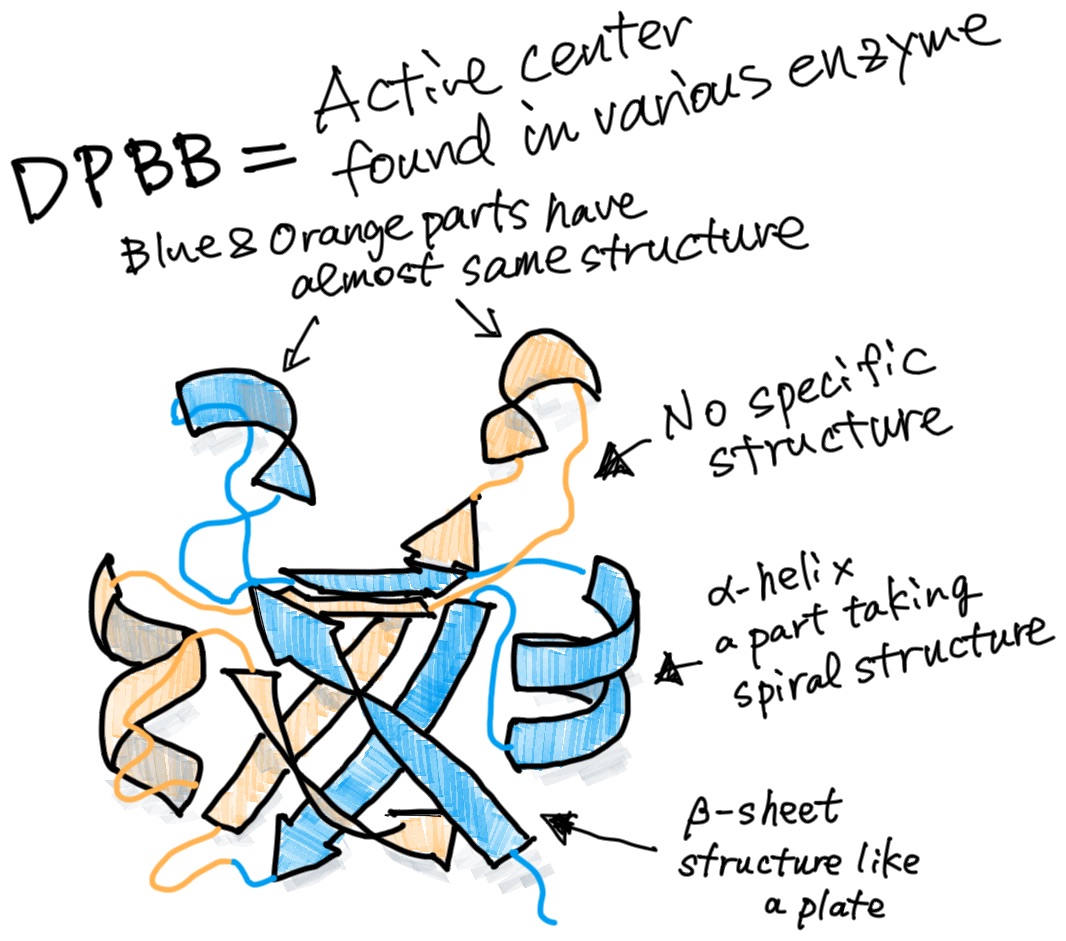
We can speculate that DPBB acquired several different functions as it evolved. So, we are trying to create an ancestral DPBB protein state.
I see.
Looking at the structure of DPBB, you can see that the structure of the N- and C-terminal halves are similar. So, it was thought that the ancestral DPBB was a dimer of these half peptides, and the current DPBB resulted from a duplication and fusion of the half gene.
So, I first decided to try making the sequence of the half-sized DPBB to check if it could fold into this structure.
When I did, the half-sized peptide really did form the DPBB structure through homo-dimerization.
Additionally, from a structural biology perspective, it was thankfully easy to crystallize for structural analysis. When we tried to crystallize some mutants under 100 conditions, we could obtain crystals under about five or six conditions.
That’s a high probability. Usually, you have to consider more than 1,000 conditions before you finally get one to crystallize. That’s quite an efficient experimental system.
That’s right (laughing). I think this protein has a relatively “rigid” structure making it easy to crystallize. It’s also stable at high temperatures, like 80℃.
In the discussions on the origin of life, there are mentions of the hydrothermal vents in the deep ocean, so this seems plausible.
Simplification of amino acid components in sequence
This protein is made up of a sequence of 43 amino acids, but seven of the 20 amino acid types are not used. In other words, it is made of 13 different amino acids. So, my next question was, “Is it possible to further reduce the number of amino acid types?”
How can you do that?
First, we reduce the number of amino acid types one by one, such as making a variant without methionine and another variant without leucine. I made six different variants, each without one type of amino acid, and to my surprise, I found that they all formed DPBB structures.
Wow. It must be a very strong structure.
Not only that, but I also tested variants without two or three types, and those also formed similar structures.
That’s amazing.
And…
(There’s more!)
In the end, I made a variant without all the six types of amino acids I was testing, but this variant did not form any structure in solution.
(Too bad…)
But…
But…?
The crystallization process was a part of my routine, so I tried crystallizing this variant as well. Surprisingly, it did crystallize. I was able to confirm that the structure was almost identical to the others.
So, you can make this structure with only seven amino acids? That’s amazing.
Primitive amino acids and primitive proteins
Actually, it’s even more interesting to look at these seven amino acids in a codon table.
By codon table, you are referring to the table showing how the DNA sequences correspond to amino acids.
Yes. Looking at this table, you can see that these seven amino acids are clustered near the bottom right side.
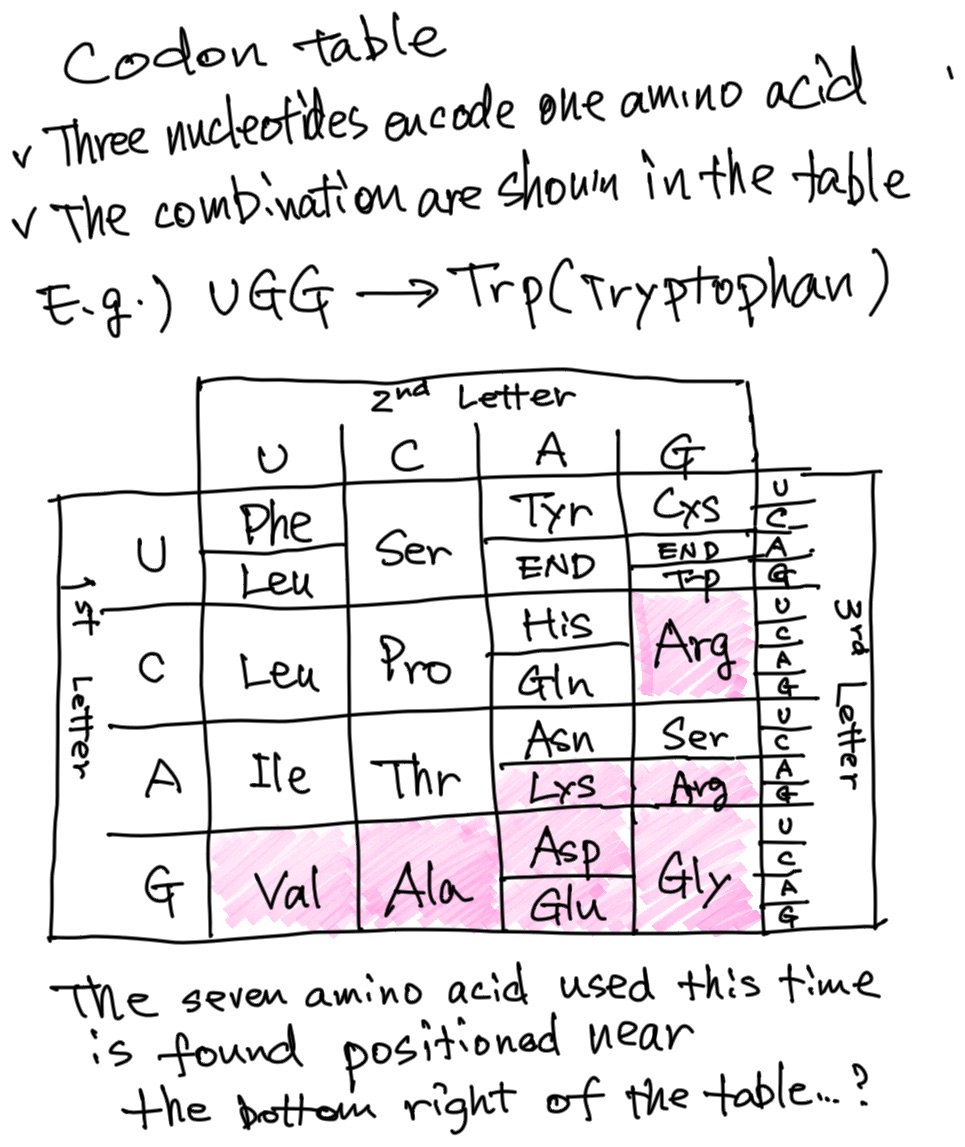
I see that.
In my recent work, I was able to reproduce the DPBB structure using only seven of the 20 amino acid types. The amino acids used in this experiment also have relatively simple molecular structures, so it’s easy to think that these amino acids also existed in the ancient world. If so, it is conceivable that DPBB could have emerged even in early Earth.
I understand how the structure could be made, but I’m curious about the function.
That is a frequently asked question.
To be honest, I don’t have the full answer yet, but we have found that it binds to DNA. β-barrel structures similar to DPBB are found in transcription factors and ribosomal proteins. So, now, I’m picturing that the ancient DPBB structure may have also bound to RNA and other nucleotides to do something.
In any case, nothing will start if they don’t bind together, and when they were bound together, by chance, it might have turned useful. This cycle was probably repeated over and over. If you think about it in terms of the Earth’s timescale, there is a hundred million years to experiment as much as you want.
It’s very difficult to figure out how proteins evolved in the early world. At worst, it’s like science fiction thinking. So, I think one solution to investigate protein evolution is to experimentally reproduce the evolutionary process. Now that I have been able to simplify RNA polymerase to this level, I would like to look at the evolution of RNA polymerase even further in the past.
Clues for researchers focusing on evolution of translation
The tools used are so advanced nowadays that it seems like a world apart from 20 years ago.
That’s true. This time, we used computational calculations to design the sequences and to predict their structures, but I couldn’t do that myself. So, we collaborated with Kam Zhang and his team in the Laboratory for Structural Bioinformatics. I have also recently been using an AI system called AlphaFold.
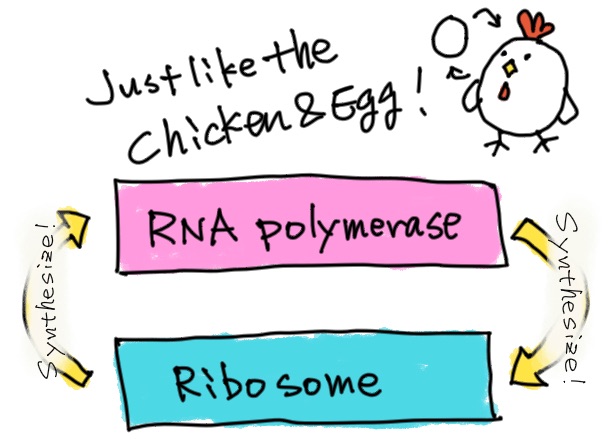
We talked earlier about the chicken and egg problem, but because RNA polymerase is also made of proteins, I guess research on ribosomes, which make proteins from RNA, is also important.
This time, we have discovered that the core of the RNA polymerase protein seems to be composed of only seven amino acids types.
Oh, I see. So, the next step would be to make a ribosome that can polymerize these seven amino acids.
That’s right. Modern proteins consist of 20 amino acid types, so we need to know what kind of function is needed to polymerize the 20 different amino acid types in the translation process. But we discovered that the core structure of RNA polymerase can be made from just seven amino acid types, so it should also be possible to reduce the number of amino acids needed to seven when considering the evolution of translation system.
It sounds like the bar has been lowered a bit.
There are a lot of people around the world doing research on ribosomes, and I think our results provide them with some clues.
Your contribution to this topic was made possible because you went against the grain and chose to study RNA polymerase, which no one was really working on.
So, I hope we can collaborate with teams working on translation in the future.
Postscript
This time, we started with the surprise fact that the “origin of life” is actually one of the fundamental themes of biology. It was a very interesting interview because of my own personal interest in the topic. It also made me realize that technology has advanced to a point where we can experimentally verify different ideas.
