2023-05-01
Research Highlights
Research highlights articles and press releases between November 2022 to April 2023
How mouse embryos determine left from right
Apr 14, 2023
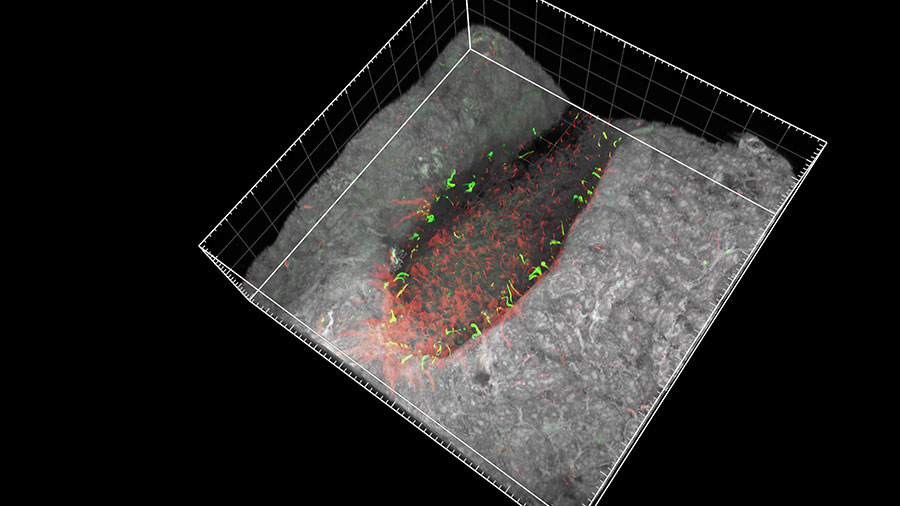
In mammals, many organs such as the heart, liver and spleen are positioned to either the left or right of the central left–right axis. During early embryonic development in mice, the motile cilia of a specialized group of cells at the node (top image) beat in a coordinated, clockwise direction, setting up a leftward flow in the surrounding fluid. This fluid flow is then picked up by static cilia, called immotile cilia, located to the left and right of the moving cilia. This detection of fluid movement causes the left and right sides of the embryo to develop differently. However, debate surrounds how the immotile cilia sense the fluid flow. Hiroshi Hamada and Takanobu Katoh (Lab for Organismal Patterning) and co-workers have put this debate to rest by showing that the cilia in mice embryos detect the movement mechanically. Using microbeads trapped in a laser beam, the team manipulated a single cilium and observed a signal involving calcium ions. This showed that mechanical stimulus of just one cilium can determine left–right asymmetry in embryos. More
Katoh TA, Omori T, Mizuno K, et al. Science 379, 66-71 (2023)
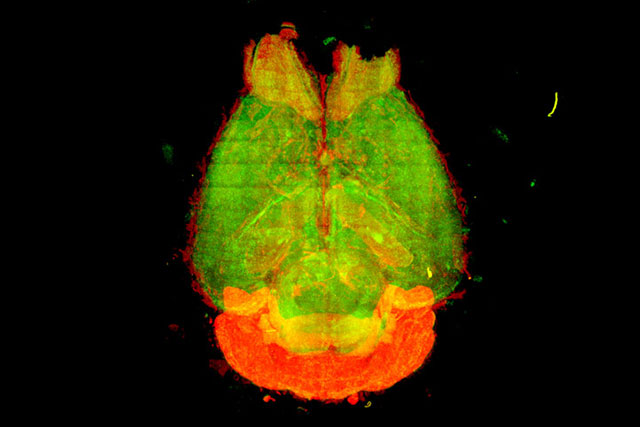
Phosphate tags on brain enzyme govern sleep dynamics in mice
Apr 6, 2023
The sleep-wake cycle is a physiological function that determines the rhythms of when we fall asleep and wake-up. Previously, BDR team leader Hiroki Ueda (Lab for Synthetic Biology) and his colleagues had identified sleep-promoting proteins whose activity could be modulated by phosphate tags. Now the researchers have shown how different phosphorylation patterns on one sleep-promoting protein—a neuronal enzyme called CaMKIIß—in mice can control different sleep induction and maintenance processes in their brains. Understanding these mechanisms may help identify new targets for the development of new therapeutic strategies for sleep disorders.More
Tone D, Ode KL, Zhang Q, et al. PLoS Biol 20, e3001813 (2022)
A nifty trick to help plants thrive in iron-poor soils
Mar 16, 2023
Iron deficiency in the soil can have significant negative effects on plant growth, yield, and overall health. Barley and wheat have evolved a unique strategy to capture iron. They release compounds called mugineic acids that are released into the soil, where they bind with iron and form a complex that the plants can absorb. The mechanism of their uptake by plants involves a specific transporter system in the root. BDR senior scientist Atsushi Yamagata (Lab for Protein Functional and Structural Biology) and his co-workers have now determined the structure of the transporter protein for the first time using cryo-electron microscopy. This research is now guiding work to develop derivatives of mugineic acids, which the team believe could become a new generation of highly effective fertilizers for alkaline soils. More
Yamagata A, Murata Y, Namba K, et al. Nat Commun 13, 7180 (2022)
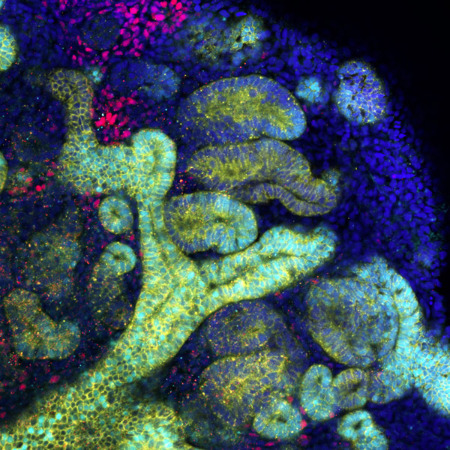
The next milestones in generating artificial organs
Dec 21, 2022
The ultimate goal of research using human pluripotent stem cells, in the context of regenerative medicine, is to recreate whole, replaceable organs in a laboratory setting. Transplanting these organoids could improve many treatment options. Organoids are developed from stem cells and cultured on 3D frameworks, where they self-organize into functional tissue. These can partially replicate what cells within certain organs do. However, the largest organoids available today are only a few millimeters across. “Once we discover how to grow them at scales comparable to real organs, we could create artificial organs derived from a patient’s own stem cells” says BDR team leader Minoru Takasato. He is involved in a consortium that brings together RIKEN researchers and external collaborators to tackle organoid challenges and find real-world applications. More
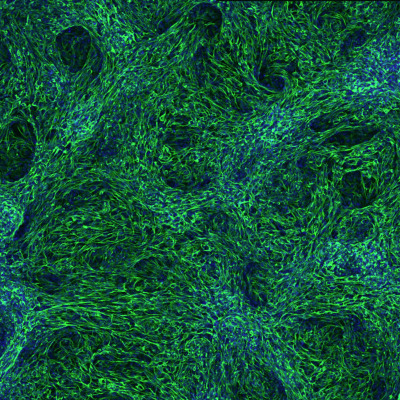
An experimental strategy for recreating organ formation in the lab
Dec 14, 2022
The visceral organs are principally formed by embryonic epithelial cells—cells that line surfaces in the body. But recent research has revealed that this process involves close communication with the mesenchymal cells that ultimately give rise to the circulatory system, skeleton and various connective tissues. BDR’s Mitsuru Morimoto (Lab for Lung Development and Regeneration) and his collaborators at Cincinnati Children’s Hospital have developed a laboratory protocol that selectively activates certain signaling pathways to convert human stem cells into various mesenchymal lineages, which support the formation of the liver, gut and other organs. In addition to shedding light on organ development, this could eventually enable doctors to provide treatments tailored to individual patients.More
Kishimoto K, Iwasawa K, Sorel A, et al. Nat Protoc 17, 2699-2719 (2022)
Inducing hibernation-like state in mice can protect organs during heart surgery
Nov 18, 2022
Researchers led by BDR’s Hidetoshi Masumoto (iPS cell-based Cardiovascular Medical Research) and Genshiro Sunagawa (Lab for Hibernation Biology) have developed a new method of protecting organs during heart and aortic surgery when blood circulation has to be blocked. Rather than relying on cold temperatures to induce hypometabolism and reduce the need for oxygen, the technique works by stimulating Q neurons in the brain, which slow metabolism down to a hibernation-like state. In this proof-of-concept study, the procedure protected mouse kidneys from damage due to lack of oxygen and avoided harmful side effects related to extended hypothermia. The findings could lead to new ways of performing similar surgeries in people. More
Kyo S, Murata K, Kawatou M, et al. JTCVS Open 12, 201-210 (2022)
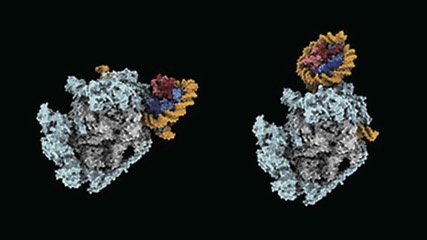
Gene-reading enzyme razes and rebuilds DNA-winding structures in its path
Nov 17, 2022
The eukaryotic DNA is tightly packed and organized into bead-on-string-like structure of chromatin–a mixture of DNA and proteins that make up chromosomes. These ‘beads’ are known as nucleosomes. Nucleosomes pose a challenge to the DNA-transcribing enzyme called RNA polymerase II, which is responsible for converting genetic information stored in DNA into messenger RNA transcripts. It has to pass through nucleosomes without altering their structure. But just how it achieves this had not been clear. Now, a team led by BDR team leader Shun-ichi Sekine (Lab for Transcription Structural Biology) has shown using cryo-electron microscopy that RNA polymerase II takes apart nucleosomes and then puts them back together again. This process allows the enzyme to access genomic sequences while ensuring that epigenetic details in the structures are not lost. More
Ehara H, Kujirai T, Shirouzu M, et al. Science 377, eabp9466 (2022)