What’s Important for Experimental Automation?
This time I talked to Dr. Atsushi Shibai, who works in a laboratory that researches experimental evolution and experimental automation. I previously talked to Dr. Uchida from the same laboratory about her research using medaka fish, but I heard that Dr. Shibai is more of an engineer-minded person… So, I went to delve deeper into who he is.
A graduate of Nara KOSEN!
Let me cut to the chase. What kind of research are you doing?
The research projects in the Furusawa lab, which I currently work in, can be largely divided into two directions; experiments involving making bacteria evolve in the laboratory and working to automate those experiments. My research focuses more on the side of automation.
So, your background is not in biology but automation?
I was actually studying robotics during my time at Nara KOSEN. The regular course at KOSEN is a 5-year curriculum, but I spent another two years after that enrolled in a specialized advanced course, meaning that I was at Nara KOSEN until I finished the equivalent of fourth year university education. I started working on bacteria research for my masters degree and I came to RIKEN after I received my PhD. All that led me to my current main work of automating experimental bacterial evolution using robotics.
What is experimental bacterial evolution?
What kind of experiments are you doing for experimental evolution of bacteria?
Have you heard of drug-resistant bacteria? “Drug resistant” means bacteria become more resistant to antibiotics. From the human perspective, this type of bacteria is troublesome because they cannot be killed with antibiotics, but from an experimental point of view, it is important to understand how these bacteria have evolved to adapt to its environment in order to survive.
In my experiments, I decide in advance the target level of resistance of the bacteria to each of the different drugs to be used, and then test whether I can steer evolution of the bacteria in a targeted direction by administering multiple drugs simultaneously. Recently, there are super drug-resistant bacteria that have emerged, where antibiotics of all kinds are proving to be ineffective. If we can reveal how they evolved into such bacteria, we may be able to find ways to use antibiotics so that bacteria are less likely to develop drug resistance.
How do you control evolution?
When you administer antibiotics to bacteria, most of course will die, but there will be some bacteria that will survive that are slightly resistant to the drug administered. You then select the bacteria that survived, administer the drug again, select the surviving bacteria, and then repeat this process over and over. Gradually the bacteria will evolve into a drug-resistant strain.
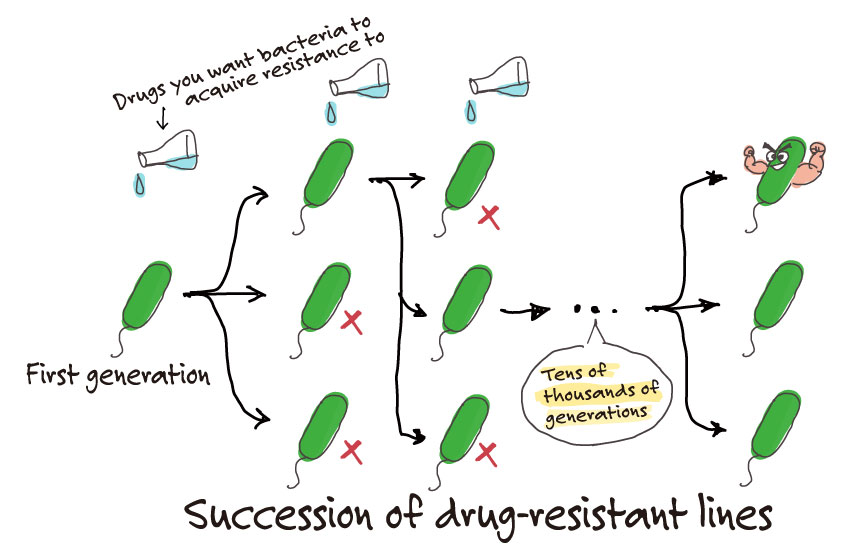 If you are looking at drug resistance to one drug substance, you can start by culturing the bacteria with low concentrations of the said drug and gradually increase its concentration. But if there are three drugs, there is an infinite number of combinations of the concentration balances of each drug. We need to determine experimentally which out of the infinite number of combinations can efficiently drive evolution of the bacteria. You would think that the most efficient evolutionary pathway would be for the bacteria to develop resistance to all three drugs at the same pace. However, if you run experiments, you find that the bacteria will first acquire resistance to one drug, and then will acquire resistance to the other two after that. This seemingly roundabout evolutionary pathway is in fact the most efficient way for bacteria to evolve and reach the targeted resistance.
If you are looking at drug resistance to one drug substance, you can start by culturing the bacteria with low concentrations of the said drug and gradually increase its concentration. But if there are three drugs, there is an infinite number of combinations of the concentration balances of each drug. We need to determine experimentally which out of the infinite number of combinations can efficiently drive evolution of the bacteria. You would think that the most efficient evolutionary pathway would be for the bacteria to develop resistance to all three drugs at the same pace. However, if you run experiments, you find that the bacteria will first acquire resistance to one drug, and then will acquire resistance to the other two after that. This seemingly roundabout evolutionary pathway is in fact the most efficient way for bacteria to evolve and reach the targeted resistance.
Automation is impressive
How long does it take to complete evolution?
It takes about 20 years for humans to generate one generation, while bacteria can divide in about 20 minutes, so that’s about 2,000 generations in one month. During that time, they divide every hour of every day, so regardless of whether its Saturday or Sunday, they need to be passaged. As I mentioned earlier, in an experimental system for bacterial evolution, we select multiple conditions from among the infinite number of drug concentration combinations, so we are always maintaining about 300 different strains. This means an enormous amount of work needs to be carried out at the same time. Furthermore, the amount of solution administered varies between strains and
from day to day, meaning we need to change the pipette settings each time.
That means that you have to rotate the dial on the micropipette about 1,000 times to complete one operation! There is no way you want to do that by hand…
This is why automation is necessary. We also need to calculate what concentration is needed in the next step based on what was used in the previous round. These calculations are needed for all strains and for all drugs.
Humans are bound to make mistakes if they were to do all of this.
I noticed that the software that came with the dispensing robot I was using for my experiments allowed users to write a program script, so I wrote a really long program that can calculate the concentration and the amount to be dispensed. Once I did that, the rest was easy and now the setting process is much less labor intensive, and I am free from having to rotate the dial 1,000 times.
Automation and robots are impressive indeed.
Automation can be more flexible if we use ingenuity
To be honest, I sometimes wonder whether automation research commonly done in biology can actually be called automation research. It is hard work to learn how to hook up the equipment you bought and then run it as laid out by the manufacturer. But, because I went to a technical college, I am more keen to work on building and developing different things.
My research topic in graduate school was looking at the relationship between bacteria and spontaneous mutations. So my work consisted of endlessly exposing bacteria to ultraviolet (UV) light. It was a perpetual repeat of feeding the bacteria, changing the medium, passaging the bacteria, and exposing them to UV light. The first year, I figured this was part of the experience, so I spent countless hours doing that. I was making 500 plates a week, and spent 12 to 13 hours a day, including weekends, doing experiments. That was my life.

However, these tasks were very simple. So, I thought I could probably automate this process, and in my second year I built a device that could do just that. All I needed was a UV lamp, and a visible LED and photodetector pair to measure the turbidity of the culture medium. The UV light cost about 10,000 to 20,000 yen, but the visible LED and photodetector pair only cost about 300 yen. For my first prototype, the case to hold the test tube was an empty gluestick casing. It was the perfect size. In the end, I made one using a 3D printer. For the controls, I used an open source single-board microcontroller. This device, something I built from parts that I bought out-of-pocket, was actually more accurate in measuring turbidity than compared to commercially available spectrophotometers.
It’s not uncommon for the automated robots and devices that are used in biological experiments to cost tens of millions of yen per unit. On top of that, there are some challenges with them as well. For example, biological shakers must continuously be moving. When there are a lot of moving parts, they can wear and break. The purpose of automating tasks is to allow handling of large numbers of samples at once, so if you have a device that can shake 100 plates at one time and that breaks down, then 100 plates of your experiments will be ruined at once. There will also be a large number of experiments that you cannot do while you wait for the equipment to be repaired.
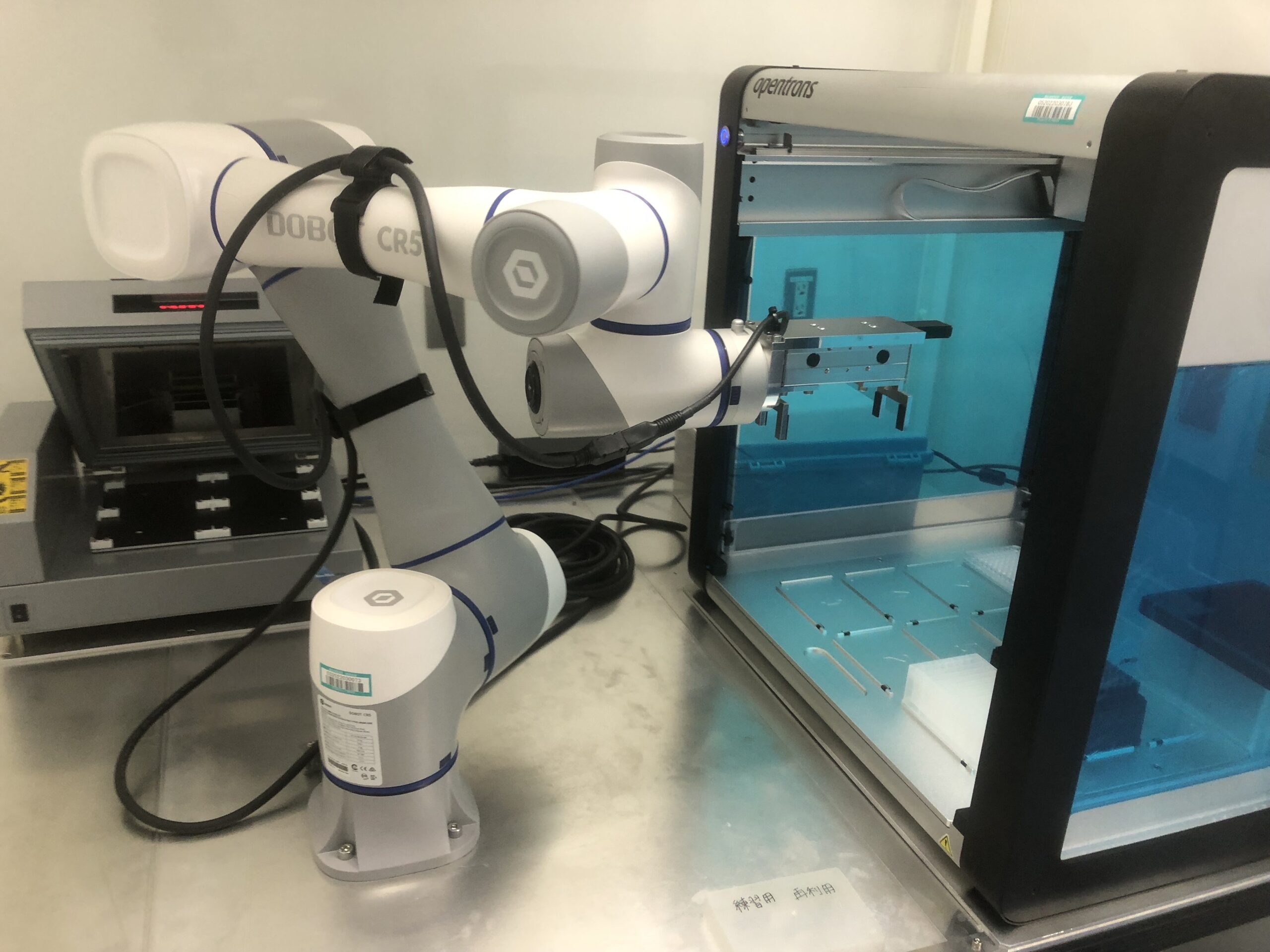
So when I automate tasks, I also build small devices. Like one where a small robot handles only a few plates. Basically, something too simple for it to break. And if you have a lot the same small devices lined up, you can handle the same large numbers of samples. I think it’s interesting to come up with these kinds of devices on your own.
Don’t rule out all options
What is it that motivates you to work in the life science field as a graduate of a technical college?
I would really like to promote the connection between biology and robots. But I also want to stay aware of human performance.
There is a national KOSEN programming contest held every year, and at a contest held about ten years ago, the participants were given a task to count the number of dice in an picture with dice of all different sizes stacked in a pile. Since it is a programming contest you would expect that the program would involve some sort of image processing. However, the first- and third-place winners had counted the dice by hand.
Isn’t that cheating?
When you read the report reviewing this contest, what was interesting was that the first-place team commented that they had tried using image processing to count the dice, but were unable to beat the performance by humans. The team that wrote the report describe the reason for their own defeat was because they were working on their program up until the last minute to improve the accuracy of one method and they didn’t have time to compare it to other methods. They admit that they should have tried counting by hand as long as the rules permit it. And it’s true that nowhere in the rules did it say “Counting by hand is not allowed.”
How you interpret this problem will affect your view on automation. As long as its not something that should not be done, we should try out all possibilities. And sometimes if we think about it very carefully, there are cases where it may not be worth automating. So I hope to maintain a flexible mindset by not going overboard with the notion that automation and robots are great, and leave open the option of humans doing some tasks.

Postscript
We touched on a wide range of topics that I had not previously heard much about during my other interviews for this series, such as stories about KOSEN and robotic competitions. We probably covered enough topics for a manuscript three times longer than this one. Our talk made me realize how different things can seem when you come from different backgrounds.
