Measuring Various Things with Mass Spectrometers
Dr. Hina Kosakamoto from whom I learned about nutrition research using fruit flies, introduced me to my next interviewee, Dr. Keiko Masuda. Dr. Masuda is an expert in mass spectrometry. Through this interview series, I’ve heard a lot about genes and cells, but this is the first time mass spectrometry has come up. I wonder what kind of research we’ll talk about today.
First step into protein research
When did you first come across protein research?
The department at Hiroshima University where I did my studies is a bit unique. Students were allowed to take half of their credits in humanities subjects and the other half in the sciences. I remember hearing the dean of the department at the time saying, “It is important to foster experts who specialize in specific areas, but we also need people who can serve as a bridge between specialists in various areas.” He also said, ”It is important to foster human resources who have developed a wide human network and also have their antennae up in different directions.”
So, your department was one that could be called “interdisciplinary.”
I always had an interest in biology, and when I was taking the university entrance exams, I had my heart set on pursuing pharmaceutical sciences. But when I was looking into Hiroshima University, I became intrigued by the degree programs of the School of Integrated Arts and Sciences. And I decided that I wanted to advance my studies with a broader perspective rather than deciding on a specific field.
Exactly what your dean was aiming for?
After studying a range of different subjects, I found that my interests still laid close to the life sciences and in my third year, I chose to major in the life science program. The laboratory that I joined in my fourth year was doing research on genes whose functions had not yet been identified. Genes are the blueprints of proteins, so we can’t understand the function of a gene until they become proteins. Thus, we were working as a team to synthesize proteins, injecting them into experimental animals and then examining their phenotypes. This was my first foray into protein research.
Common occurrences with protein synthesis
Finding out DNA sequences can be done fairly easy, but when it comes to actually making proteins, it is quite difficult. We were struggling to obtain a specific protein. This protein was about 80 to 88 amino acids long and was highly hydrophobic. It also contained post-translational modifications such as disulfide bonds, making the protein hard to purify and isolate.
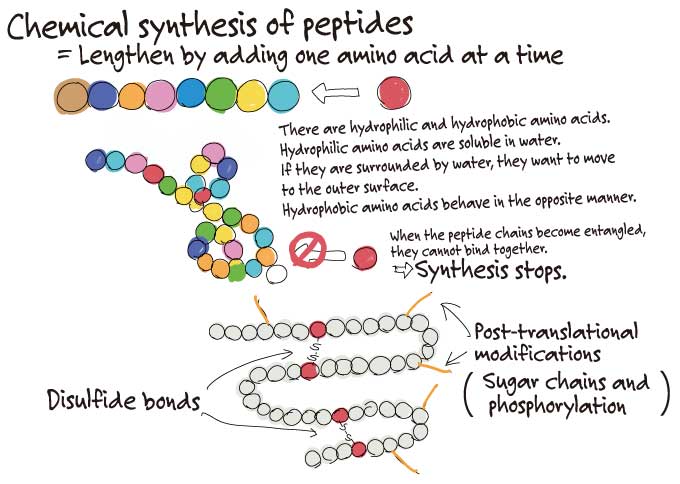
That’s a common occurrence when looking at protein expression. But if you can’t obtain the proteins, you can’t start your experiments…
Exactly. We tried to get E. coli to express the protein, but the yield was low. It was difficult to artificially synthesize the full length of the peptide using conventional chemical synthesis, so we even tried requesting a company that offered protein synthesis services to make the protein for us.
I imagine that because the peptide is pretty long and you also need a fair amount of the protein, it can become a bit costly.
The quote was apparently a few million Japanese yen. Because we needed to synthesize the protein continuously, not just once, we abandoned the idea to outsource the protein synthesis part. Instead, we ended up installing a protein synthesis machine that uses microwaves. The machine allowed us to apply microwaves to the proteins that tended to aggregate during the synthesis process to stretch the protein as it was being made and allowed us to obtain the amount of protein we needed.
Were you able to confirm the phenotype in the end?
Yes. We found when overexpressed, it can lead to fat accumulation.
Something that we all don’t like.
Development of mass spectrometry techniques
Is that when you were first introduced to mass spectrometry?
Yes. But in the beginning, I was not actually doing it myself; rather, I was asking a technical specialist to do the analysis using the mass spectrometer at the university’s common facilities. It was not until after moving to RIKEN that I began to seriously work with mass spectrometry, and it was Dr. Tsutomu Masujima (deceased) who led me to this path. It’s because of this connection that I now work with mass spectrometry and cell-free protein synthesis.
I think mass spectrometry is often used to identify substances. But it can also be used to quantify substances, right?
Our team developed a technique called MS-QBiC, which we are currently working to improve. It’s a method that involves using a peptide with the same sequence as the sample, but that is labeled with a stable isotope as an internal standard before measuring the sample with the mass spectrometer.
For example, the molecular weight of nitrogen is normally 14, but if we use the nitrogen 15 isotope, the molecular weight shifts by one. If you add a fixed amount of this labeled peptide, we can measure the amount of the target peptide.
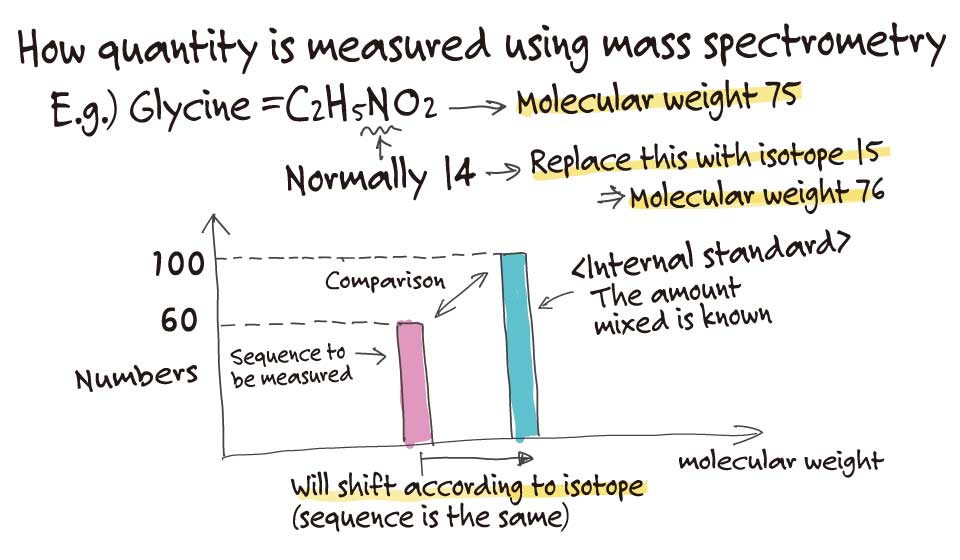
So, you are basically mixing something like a ruler into your sample. But I imagine that using stable isotopes can be quite costly.
MS-QBiC can actually resolve this issue. It is based on a reconstituted cell-free protein synthesis system, called the PURE system. This system allows us to make very small amounts of proteins. The cost of the ruler is basically the cost of the stable isotope, so if you only need a small amount, the cost of the experiment drops accordingly.
You can measure many peptides at once using mass spectrometry. Does that mean that you need a lot of rulers as well?
We’ve solved that issue as well. MS-QBiC includes a tag sequence, and if you make a lot of variations of the tag sequence, you can still distinguish between peptides even if the sample is a mix of different peptides. We were also able to improve the measurement throughput, and we are now working beyond that. Our next goal is to be able to measure post-transcriptional modifications to proteins, such as phosphorylation, which is important for regulating protein activity. It’s still in the developmental stages, however, we would like to be able to synthesize peptides with post-transcriptional modifications by modifying the enzymes and genetic codes used in the PURE system.
And that is why you are currently working on mass spectrometry and cell-free protein synthesis.
It’s fun to be able to work on different themes
Do you have a lot of research collaborations where you identify and quantify proteins?
Yes. It’s fun to be involved with a lot of different research topics. There is a lot of data that you can acquire only by using mass spectrometry. Dr. Kosakamoto, who introduced me to you, is also one of my research collaborators.
I see.
I can operate a mass spectrometer, but what is ultimately important is knowing what you want to measure. Nothing will emerge from me alone, but the conversations I have with different researchers can lead us to measure something where we think mass spectrometry might prove useful. Of course, I am happy when we get the results we wanted, but even if the results are not what we expected, there is something new to be learned from that as well. While I may not be focusing and delving into a specific research theme, I am happy and also enjoy being able to help out with a variety of research. There are many researchers around me who have a wide range of interests, and so I find that each new round of discussions, data acquisition, and then getting feedback on the data from those researchers is refreshing as I am learning something new.
It’s nice that there are also many types of researchers.
Mass spectrometry is very sensitive, and there is a lot of information that you can extract from it. People sometimes think that we just push a button and voila! the machine will spit out the results. But it is a job that requires knowledge and experience in order to optimize the workflow to produce the type of results that meets the aim for a particular project. It can be an intricate process. For example, depending on the sample, the sample preparation process or figuring out the measurement parameters may turn out to be unexpectedly complex. Or how the data that is obtained is analyzed may be important. This is what makes it both challenging and interesting.
Mass spectrometers are vacuum systems, so their maintenance must be difficult.
To ensure that we can collect good data at any time, we clean or replace parts at appropriate times and also run quality control measurements to check that there are no irregularities. To maintain the vacuum inside the instrument, we basically leave the power on. It is a real concern when there are planned power outages or when typhoons are approaching. We don’t want to turn the power off unnecessarily as it can place a heavy load on the instrument when you release the high-vacuum pressure and then reinstate it again. Repair work is costly if the machine breaks down and it’s not unusual to have to wait several months for some parts.
I guess the analogy, ‘The more troublesome a child is the more endearing they become,’ fits perfectly in this case.

Postscript
During the interview, it felt like the energy she was projecting screamed, “I love mass spectrometry!” When I mentioned this to her, she told me that was not her intention. Mass spectrometers are sometimes shortened and called “mass spec” in English or massu in Japanese in the industry. During the interview, Dr. Masuda often referred to the instrument as massu but because it maybe difficult to understand, I decided to use the full term “mass spectrometer” in this piece. I also recalled the sadness I felt in my ownpast experience when the turbo pump or some other part of the vacuum systems broke down…