Behind the Scenes of Cutting-edge Research
Today’s guest is Dr. Mika Yoshimura. She’s an expert on bioinformatics who analyzes a huge amount of data from a NGS (Next-Generation Sequencing). Her role does not usually get a lot of attention but as a woman who works in the male dominated field of information systems, she sparks my curiosity more than usual…
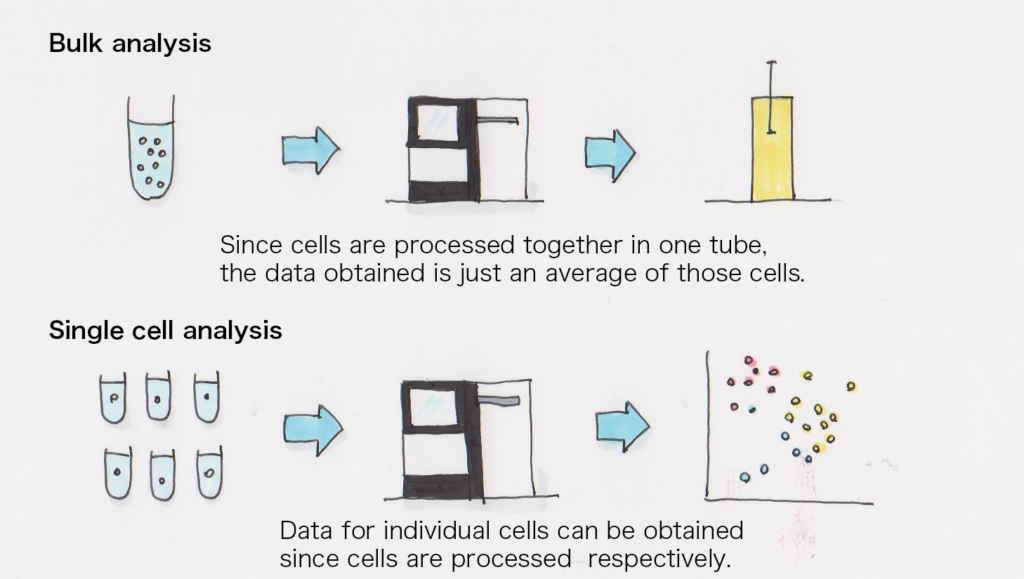
Single cell analysis
So jumping right in, based on the fact that you belong to the Laboratory for Bioinformatics Research, would you say that you prefer to work in a “dry” lab environment doing bioinformatics such as data analysis, rather than in a “wet” lab doing experiments?
Yeah, that’s true that I don’t do any “wet” lab experiments at all. My team mainly works on developing the technology for high-throughput single cell RNA sequencing (single cell RNA-seq). This is the technology that analyzes RNA transcribed from DNA in each single cell using a device called a next-generation sequencer. Our lab often collaborates with other labs using this technology and my role is to analyze the data based on the joint effort.
Of course, there are dry lab researchers with their own research themes who develop algorithms and analytical methods. Although my role is within the “dry” lab environment, I find myself supporting the “wet” lab researchers quite often.
There seems to be a lot of collaborative research, why do you think single cell is such a hot topic right now?
I think it’s because it makes it easier to understand a cell population when each of them is already analyzed individually.
In the past, the bulk RNA-seq was the mainstream method in which all cells were processed together for expression analysis, but by using single cell RNA-seq, the characteristics of individual cells in a cell population can be analyzed. For instance, we can even observe the difference between the state of each cell during the process where iPS cells become another type of cell.
In addition to that, in my opinion, by expressing the data in a matrix format, mathematical formulae can be used to analyze them which, in turn, allows us to examine them from various angles.
I didn’t think research was for me
Have you always done this kind of work before you came to RIKEN?
In both undergraduate and graduate school, I was doing research in a “wet” lab. Even my PhD thesis was based on the data I collected from the experiments that I conducted myself, so I wasn’t familiar with computers at all. But for some reason, I got a job at an IT company after graduate school.
Oh? Why? (laughing)
There were various reasons, but one of the main reasons is that I didn’t think research was for me.
First of all, I got a job as a fresh graduate at a system integrator company and then I moved to a software package development company and finally came to RIKEN.
That’s a great career. What made you decide to return to academia?
There were various reasons again. The job at the software package development company was pretty demanding and it affected my health. There is a trend in the IT industry to switch jobs in one’s mid-thirties. Since I couldn’t see a long-term career vision at that company, I wondered what to do. I found an internship at a company in the scientific education field. While I was there, I became acquainted with researchers from RIKEN and heard that they were hiring not only researchers but also engineers with a science background. That position was at the laboratory where I currently work.
Does that mean that you didn’t have any experience or skills in bioinformatics at that point?
That’s right. I didn’t have any. (laughing) I was initially hired to set up a platform for data analysis. They needed someone who has some degree of understanding of biology and coding skills, and I fit that description. I was also interested in data analysis, so now I work on both the infrastructure for data analysis and the data analysis itself.
The lab has an infrastructure team that sets up and maintains hardware. I don’t know a lot about infrastructure, but the people in the infrastructure team are highly skilled in systems engineering. The platform for which I’m responsible lays on top of what they built, and is a bit more customized for the needs of the researchers.
Various types of infrastructure
How would you describe the infrastructure?
My lab has an infrastructure called a computational cluster which is a group of servers in parallel to perform complex calculations at once, and we usually share it with our collaborators. We are currently preparing an analysis environment in the cloud, and I believe we will eventually move everything there.
Does the data analysis platform or infrastructure require more machine power for single cell analysis than bulk cell analysis?
It’s technically possible to use a standard desktop machine for analysis but it would need a specific environment for single cell analysis data computation. Calculations themselves could take literally forever without adequate amount of memory, or it could produce a few terabytes of data so it would require a large amount of storage or a group of machines that can do calculations in parallel. It’s simply not realistic to do the analysis in a reasonable amount of time to do the research.
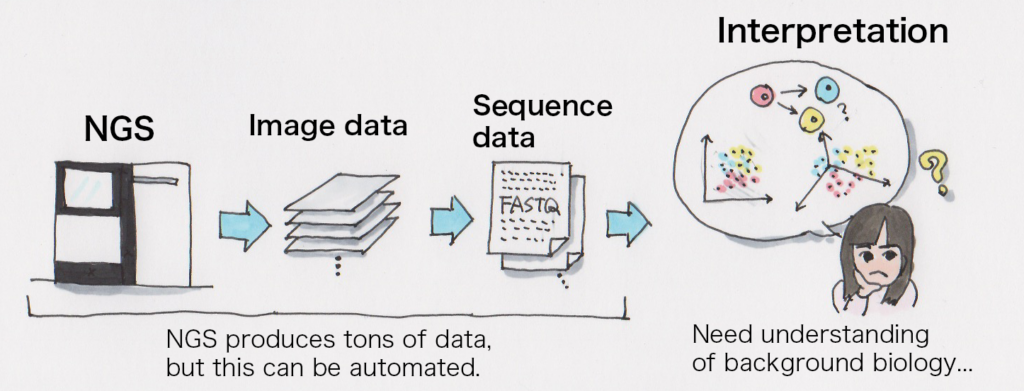
It would be a problem if analysis takes 76 years to do.
I agree. Biology and computers cannot be separated today.
Currently, my main project in the infrastructure is to build a workflow that can execute various analytical steps with a one-line command.
So basically, I don’t generally work on the hardware.
I see. That’s how you became the point of contact for the researchers
I think it’s important that servers are maintained by someone who has the specific knowledge in the field. I’ve heard that even a professor is doing such jobs at some universities. In that respect, researchers in “dry” labs are in a very privileged environment.
That’s why joint research is so common.
Exactly. I really hope that the collaborators are happy with the work I do…
Working with cutting-edge technology can be challenging
Are you worried if they’re happy with you? Why?
I don’t think I have the same depth of background and foundational knowledge of data analysis that would allow me to think outside the box like industry experts. I know sometimes discoveries are made in a eureka moment in the researcher’s mind based on their experience and knowledge. I’m not confident that my level of knowledge would lead me to that kind of discovery.
I usually consult with my boss and the researchers in the lab so that I can receive a thorough review before submitting the results.
It may be difficult to analyze without some knowledge of the biological phenomenon.
There are various cases. It is relatively smooth when I’m verifying the hypotheses that are already fairly well established. On the other hand, it can be difficult in the case of finding something new by looking at the results from single cell analysis. I try to slice the data in a number of ways, but I still get feedback from “wet” lab researchers where they point out what’s missing from their point of view. Then I go back to analyzing the same data, this time, based on their feedback. It’s a process of trial and error.
When seeking new findings, it sounds difficult to judge whether the analysis result is good or bad.
That’s right. Also, there is always a new analytics tool coming out, so I feel pressured to keep up to date, otherwise I’m afraid we won’t be able to produce good results. I constantly feel that there is a more efficient way than what I’m doing right now.
Also, it’s natural but it’s still tough to be one of a very few people who do this kind of work.
I see, that means that job is less likely to get standardized.
We need to keep updating…
Well, technology is advancing rapidly. “Wet” lab researchers are quite smart so they are capable of learning some level of coding skills. Some of them can write codes that can analyze the data to a certain degree. Currently, we have a package that is close to becoming the global standard for single cell RNA-seq analysis, which is written in R that is often used in the field of statistical processing. While R is used for economics, it allows us to create various charts that are useful for our research.
R. I’ll check it out later.
R makes it easier to draw graphs and diagrams.
R doesn’t require much programming knowledge to do a little analysis or drawing charts. In fact, there was a researcher who was able to start writing in R on their own while we were working together.
I see, then you have to keep ahead of the game… But how did you learn that at first?
There weren’t many books available back then, so I studied the code written in R by other researchers. I could follow most of it because I was trying to do almost the same thing with my code. When I didn’t understand the algorithm in their code, I read the technical books.
Your brain seems to be wired like an engineer’s…
It’s like learning by reading the code. There are many good books now.
In fact, we accept students as interns. When we train them, we ask them to read the books as well as to actually write it for themselves.
I think it would be nice if you could do a workshop on that. It would be fun.
I once hosted a training session with other researchers where “wet” lab researchers could feel comfortable participating. I’d like to do that again when I have more time.
I’m looking forward to it!
postscript
This interview felt to me as if I was getting a look behind the scenes of the exciting world of single cell analysis. I’m sure there are many things I didn’t quite understand but I feel a renewed appreciation for the people who support such cutting-edge research even though they are not in the spotlight.
