In between Technology and Science
The person I interviewed this time was Dr. Takashi Niwa, who is a member of the Laboratory for Chemical Biology. I assumed his research is chemistry oriented, but at BDR where a lot of the research centers around biology, I wondered what he actually did. So I decided to talk to him to appease my curiosity.
What chemistry can do for Biology
The name of your lab is “Laboratory for Chemical Biology”, but what exactly is it that your team is working on?
Basically, our mission as a team is to create molecules to study particular proteins in our body. My basic love is chemistry, but as the Center is mostly focused on biological research, my focus is basically on what chemistry can do for biology. When we think about it from this angle, it means developing methodologies and molecular tools.
Does that mean that you are someone who develops tools?
The Center has a large emphasis on biology in general, so we work on developing tools and methods to aid biological studies.
I’ve realized, after talking to many of the biologists here, that they often face many struggles that fall within the scope of chemistry. Since I interact with many biologists on a daily basis, I thought I could find a way for chemistry to aid the advancement of biological studies.
I suppose the biologists would like to have some kind of probe for tracking a molecule of interest?
It’s basically a probe, but more precisely, it’s transforming a molecule within a living organism into something like a probe or labeling specific proteins with a tiny molecule which can help us to visualize them. If we are successful in these developments, we can use them in diagnostics and treatments.
But that would just end up in the scope of engineering. My personal passion is still chemistry, so while I’m creating the probes, I’m also thinking of ways to design new molecules.
Human brain over computer
How do you go about designing a molecule?
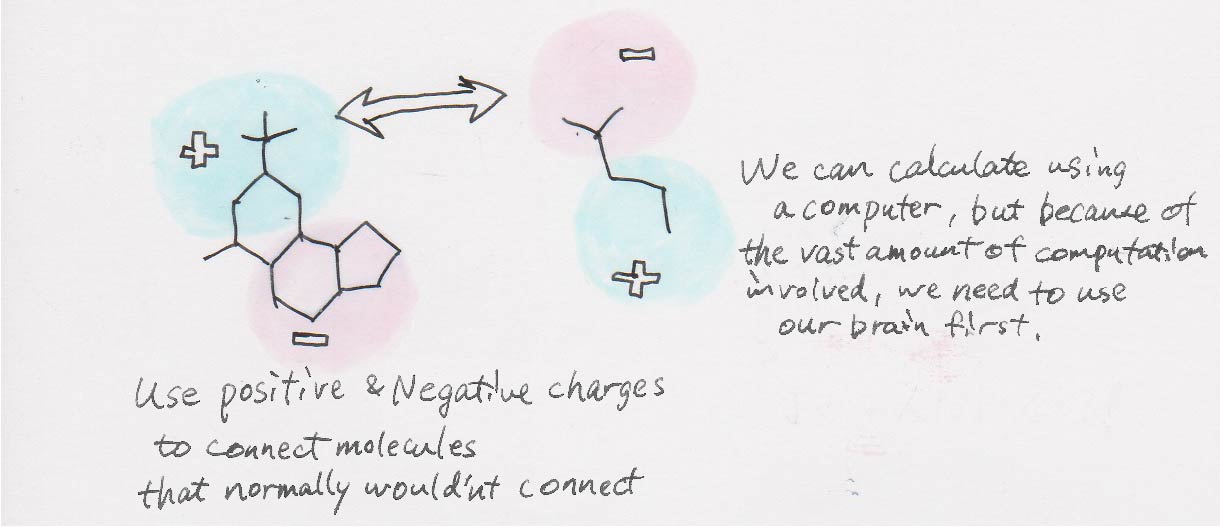
I’ve been studying the development of new molecular reactions since I was an undergrad. It’s an area in chemistry where a Nobel Prize is awarded about once every five years or so. Developing organic reactions means finding a way to link molecules together that don’t normally attach to each other.
How do you attach the molecules that don’t normally attach to each other?
There are many ways of doing this but the simplest way is to basically use their electrostatic properties, where negatives and positives attract each other. It’s similar to the way that sodium ions (Na+) and chloride ions (Cl–) can make sodium chloride (NaCl) because of their positive and negative charges. When you take a closer look at organic compounds, we can see that there are fluctuations in the electric charge distribution. Organic chemistry starts by linking the chains that we suspect have negative and positive charges.
I can imagine that if it’s a small molecule, but if it’s a big molecule, would you be using simulations?
Absolutely. Based on my experience and knowledge from reading a number of textbooks and research papers, I can usually tell which part might be positive or negative from looking at the shape of the molecule. But in order to have a rigorous discussion, we can’t just rely on our instincts, so we use supercomputers to simulate reactions.
Does that mean nowadays you can design molecules using simulation?
Yes and no. There are unlimited ways that chemical reactions can happen. The molecule only settles to a stable state after first crossing the energy barrier, but there are countless paths to crossing the activation energy barrier. This means that it’ll require too much time to calculate even using supercomputers.
Furthermore, even if we think linking A and B will create C, we also have to consider the possibility that A is really reacting to something other than B to create D, and that D and B are reacting to create C. If we try to calculate all of these possibilities using a supercomputer, the calculations will take way too long so we have to use our brains to narrow it down to more plausible scenarios before starting the simulations.
I’ve seen the diagrams depicting molecules with electron clouds. Is that an appropriate image of how you design the molecules?
Those diagrams are actually rendered after a successful reaction to understand why it happened.
That’s interesting…
Easily transforming molecules
If you get a request from a biologist to help them look at something specific, how do you go by designing a molecule?
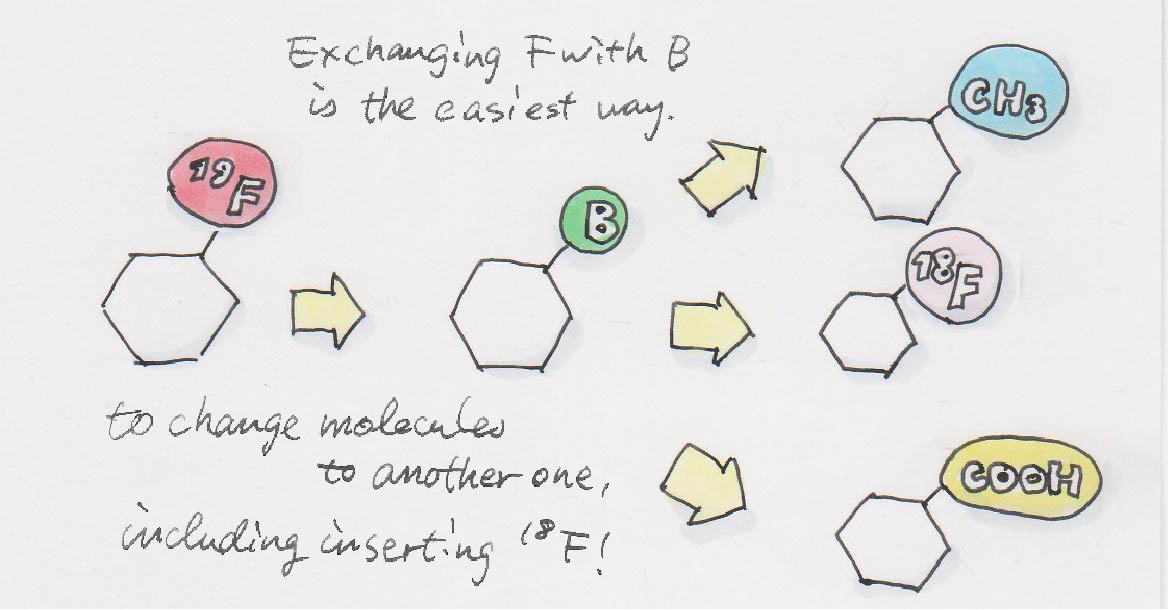
If there is a molecule that I can use as a model, I just use that. An instrument called PET, which is often used in diagnosing cancers, allows us to track the molecules of interest inside the body. However, using PET to trace the molecule requires the incorporation of a radioisotope into the original molecule.
You mean something like 18F (radioisotope of fluorine)?
Yes, exactly. Fluorine is often used to stabilize molecules in pharmaceuticals. It’s efficient if we can substitute fluorine which is normally not radioactive normally to one that is radioactive.
Are you saying that you remove the original fluorine (F) and then attach a radioactive fluorine (F*)?
Exactly. What we actually do is to first exchange the F to a boron (B), and then change the B to an F*.
The nice thing about this approach is that the F* of the end product can be anything. Removing an F is a cumbersome process, but B can be removed relatively effortlessly. Thus, we can construct various molecules based on the structure of the original molecule. We call this “molecular renovation.”
It helps to share failed results
What do you do if there is no “model” molecule?
Well, even if there is a model molecule, we can’t always use it. Sometimes there is nowhere to insert an F or there is no way to insert a carbon (C) even with the currently available technology. This means we can’t label those molecules.
When there is no model molecule, we need to begin by ensuring the person making the request can wait for six months to a year. Even then, there are some cases where we just could not design a successful reaction pathway.
Does that mean that that work will never be published?
Yes, but those are not the only instances of failure. Even if we spend six months or a year synthesizing a probe, we don’t know if it is a success until we actually inject it into a living organism. Sometimes, the molecule gets metabolized as soon as it is injected into a living organism and we can’t see anything.
That seems like a wasted effort…
Living organisms are complex so there is not much we can do.
That’s also why the “molecule renovation” approach started gaining momentum because it developed from an idea to make this molecule design process more efficient.
I still think it’s a shame that all of the data from failed experiments are lost…
I agree that there’s a lack of information on failed experiments. I also agree that emotionally it can be hard for researchers to share the data from their failures, but there is value in knowing about these cases. There are actually some scientific journals that collect these kinds of data.
I imagine that it’ll speed up the process significantly if you can just look up the experiments that failed without having to try it out yourself.
If we assume that someone else has tried out the same thing that I’m thinking of, then we can look it up somewhere and find out the details and the data. That’d be more efficient.
The failed cases would be useful, but successful cases are sometimes difficult to evaluate.
People who work on organic synthesis are often called craftsmen, and they hone their skills as they undertake undergraduate, masters, and doctoral research , but their proficiency can vary..
The world of craftsmen and trace impurities…
Do you mean that the collected data can be dramatically different for the same reaction?
Yes, it’s actually true. There are cases when it works for one person and doesn’t work for another. The reason for that is that there is still know-how missing in the procedures for an experiment, no matter how detailed we try to write them.
You mean like how you shake the test tube would make a difference? (Laughing)
It’s literally like things like that. Things like the speed at which you add a reagent, the amount of solvent used in the purifying process, the amount of water or the method of purification when a column is used can make a significant difference. Also the differences in people’s instincts as well as methods between labs can make a difference.
In fact, when I published a report on the borylation reaction to remove the F that I was referring to earlier, I got emails asking me why they couldn’t reproduce it in their labs.
I’m surprised to hear that there are situations where another lab can’t reproduce the same results in the realm of chemistry.
I’m always worried about whether my reactions are reproducible, so I generally ask my lab members to do the same experiments to ensure that the results are reproducible at least in our lab. I know that some labs are able to reproduce the reactions but there are others who can’t. The differences between those labs could just be the size of the container, the container used when heating, the purity of the reagent, etc. It really could be anything, so we usually don’t know until we go and take a look in person.
Do you mean purity of the reagent? That reminds me of when I used to work at a reagent company and we received complaints about chemical reactions not working when a different production lot was used.
That doesn’t surprise me at all. We use certain manufacturers for certain compounds as well. Changing them could potentially cause unnecessary headaches for us.
There is this well-known story that happened in the early 2000s. The metals used as catalysts can be quite expensive, but there were multiple publications that came out reporting that iron can be used instead. Then a few years after that another report was published which contradicted the earlier publications, showing that reactions can’t be observed when using high-purity iron. In the end, it was determined that the copper found in ppm order in the iron, was acting as the catalyst.
That means the purity of the iron was 99.999%. Are you saying that the less than 0.0001% of copper found in the iron was reacting?
We’ve experienced something similar in our lab, too. We were trying to convert boron to a cyano group with a radioactive carbon (11C) attached, but we just couldn’t achieve the radioactivity that we were expecting. We thought that the normal, non-radioactive carbon (12C) of methane gas could be the reason for this. But, we later ruled it out because we use an air-tight reactor called a “hot cell” for the synthesis reactions so there is no way for methane gas to get mixed in.
We then investigated whether purity of the elements could be a cause, and found out the culprit was ammonia gas. Ammonia gas is a source material used to produce the 11C-cyano group and its purity was 99.999%, which is not bad at all. But we suspected that a minuscule amount of 12C-methane gas may be mixed in, so we switched to using “ultra-high purity” ammonia which has a purity of 99.9999% and the problem was solved.
It may sound simple, but 99.999% purity is still categorized as “high-purity” so it took us a while to figure out what to look into.
We actually don’t know what’s really happening
I pictured chemistry to be more simple and sophisticated, but it sounds like you can be thrown a lot of curveballs.
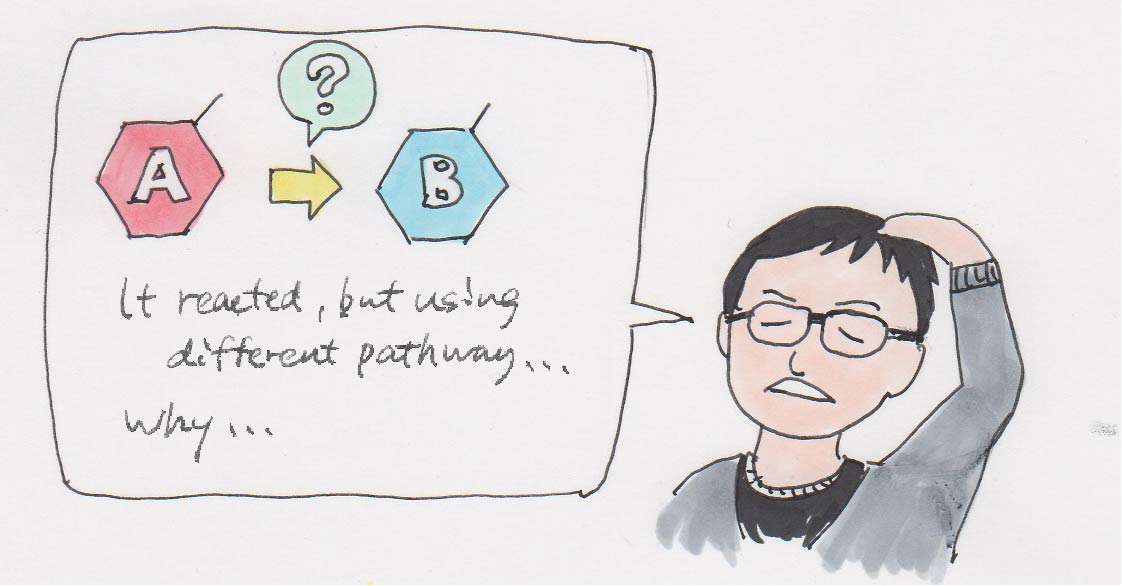
Definitely. “Molecule renovation” sounds cool, but in reality, we often don’t know why the reaction was successful. I mentioned earlier that the electron distribution diagram is rendered after the reaction is completed, but often the reaction pathway is different from what we had anticipated.
Oh, is that so?
When we are developing reaction pathways, it’s basically engineering rather than doing science. I think that it becomes science when we go beyond that scope of why an experiment was successful. I enjoy doing both, but I must admit that there are some experiments that haven’t reached the realms of science yet.
For example, we use nickel for certain reactions based on past experience and publications. While there are a number of scientific explanations, it’s hard to describe it in layman’s terms or difficult to evaluate quantitatively as we partly rely on our instincts. I’ve been told it’s like I have the instinct of a craftsman when they asked me why I used nickel.
So it’s like you have to elucidate the reason behind the successful reaction.
On one hand, it’s sufficient to just say fluorine was converted to boron, but from a scientific perspective, we are still lacking in explanations. It’d still make a great thesis though.
I imagine that you will keep on developing many things since you’ll be involved in biological research?
Yes. Since I’m already involved in biological research, I’d like to continue contributing to molecule transformations that can be useful for biology. But again, this is more an engineering approach, rather than science.
What, then, is science I wonder? (Laughing)

Postscript
I hear a lot about the difficulties of producing the results in biology, but I didn’t realize the same is true in chemistry. I was amazed to hear that the way a flask is shaken or the way the reagent is added can make a difference in synthetic efficiency. I’m still dumbstruck by the fact that purity of 99.999% wasn’t considered pure enough and they had to use higher grade of purity of 99.9999%.
