Computational work to see the structure of DNA
今回は英語でのインタビューになったので、全編英語の記事です。でも、できる限り簡単な英語を目指したので、ぜひ挑戦してみてください。
The interviewee this time is Vipin Kumar, a former Research Associate in the Laboratory for Cell Systems Control. Because he is a non-Japanese speaker, I had to conduct the interview as well as write up this interview in English. I had an opportunity to casually chat with him the other day about much broader topics including politics, religion and education. But, I wondered how this interview would go…
*This interview took place in October 2019.
Playing with numbers
Which lab do you belong to?
I’m a member of the Taniguchi lab. I mostly do computer work developing tools.
Tools?
Yes. We are dealing with a lot of flow of numbers and measurements from the microscopes, sequencers, or whatever instrument a particular person is using. Scientific instruments generate lots of numbers and measurements. They are all numbers and it’s often not obvious how to interpret the data. I have recently been working more with sequencing.
To see what?
We directly measure the structure of the DNA. DNA is like a fiber that is two meters long. And you need to pack it inside the nucleus, which means it needs to be folded up like Origami.
We are trying to describe how it can be folded in a proper way.
It sounds hard to put something two meters long into a small space like a cell. Ah, it’s not the cell, but the nucleus!
Yes, DNA is found inside the nucleus. There is a tremendous amount of compaction, which requires a very specific kind of folding that allows DNA to fit inside the nucleus. But at the same time you need some parts of the DNA to be accessible.
Since it needs to be transcribed?
That’s right! We also need to know how to make DNA active so that it can be ready to be transcribed by specific proteins like RNA polymerases. This particular trade-off means that the cell has a unique folding mechanism, which is what we are trying to show.
I think there are several types of interactions involved in transcription, right?
True. The technology we are using allows us to see those specific interactions with the rest of the DNA.
Even long-distance interactions among different parts of DNA?
Absolutely. People often mention the promoter and enhancer regions. Sometimes those interactions are occurring between regions that are separated by long distances. This means that compaction of DNA needs to ensure these long-distance interactions can take place in addition to making DNA fit inside the nucleus.
How can we see it?
The first thing we need to do is DNA fixation using formaldehyde or some other fixing chemical. Once fixed, there will be many regions within the DNA that are glued together, i.e. interacting. Then, we cut them.
Randomly?
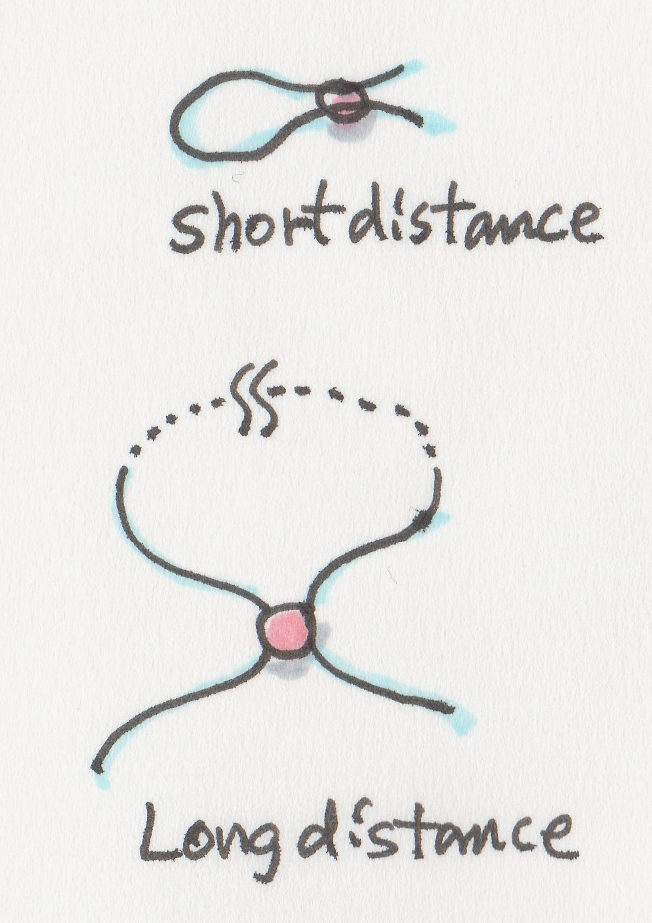
We use a specific enzyme that hopefully cuts the DNA as evenly as possible so that we cover the whole genome uniformly. The following ligation step will join together the DNA regions that were ‘glued’ to each other because of their spatial proximity, generating hybrid ligation products. We call them hybrids since they are ligated together because of their spatial proximity rather than their genome coordinate proximity. We are therefore now capturing both long and short range interactions through these hybrid ligation products.
And then?
Those pieces are sequenced using Next Generation Sequencing (NGS). Once we obtain the sequences, those DNA fragments will be mapped onto a reference sequence of the genome. This allows us to determine which regions of the DNA are interacting.
This is the Hi-C (technique), I guess?
Exactly! We are also using other techniques to see other specific interactions, too.
Heterogeneity
But these kinds of interactions should differ from cell to cell.
True, and that’s why single cell analysis is important. Basically, we can observe individual cells one by one under microscopes, but sequencing does not allow us to do so. Some microscopy techniques allow us to see the structure of DNA inside the nucleus. You will see some hubs inside the nucleus. Those hubs are the active sites, and the pattern is different from one cell to another.
The important technical aspect of Hi-C is that it involves using sequencing data from a pool of many, many cells to tell us what the average structure of the chromosome is. But as it is an average of many individual cells, it is a bit abstract and may be giving a very different picture to what is actually found in individual cells.
I see.
I’ve seen some recent results from single cell analyses that show we need to be careful about the way to look at the cells, and that there’s a huge range of heterogeneity between individual cells, which is why we don’t often observe the same interactions.
Can we do Hi-C at single-cell resolution?
Hi-C is a good tool, but its resolution is not enough to see single-cell levels. There are still many technical challenges.
But, we did recently reveal the global range of structure of the DNA.
Ah, I read about it! It was awesome.
Research highlight ‘Finding more chromosome structures by assuming less’
Press Release (in Japanese):
Science means to challenge oneself
I think I should have asked you this first, but I have some basic questions for you, like “where are you from?”
I was born in Brussels, Belgium. I was fortunate that my parents gave me a good education and went to the University of Cambridge in the U.K. for my undergraduate degree. Afterwards, I went to the University of Copenhagen in Denmark to do my master’s degree and then moved here to do my Ph.D. as part of a joint Ph.D. program with the University of Copenhagen.
How long have you been in Japan now?
This is the end of my fifth year in Japan.
Just five years? What made you decide to come to Japan?
When I finished my masters, I was very interested in stochastic systems and in particular gene expression.. This was one of the themes in the Taniguchi lab at that time and right now as well, and was a perfect fit to what I wanted to do. But later on, my doctoral work led me to doing something different, so I had to make a big change to my research theme.
Through some interviews that I’ve done, I find that many researchers currently have different research topics from what they did during their doctoral thesis.
Maybe because doctoral work is a lonely place.
A lonely place?
Yes. Most of the time, you have to defend your thesis by yourself. You need to have a strong stance to do so, but there is also the realization that maybe your thoughts are wrong. Like, what you present in the thesis is not real because you started from a wrong assumption.
You all might think that scientists are carrying out research in a very linear way. But it is not like that at all. It’s more like a spiraling path.
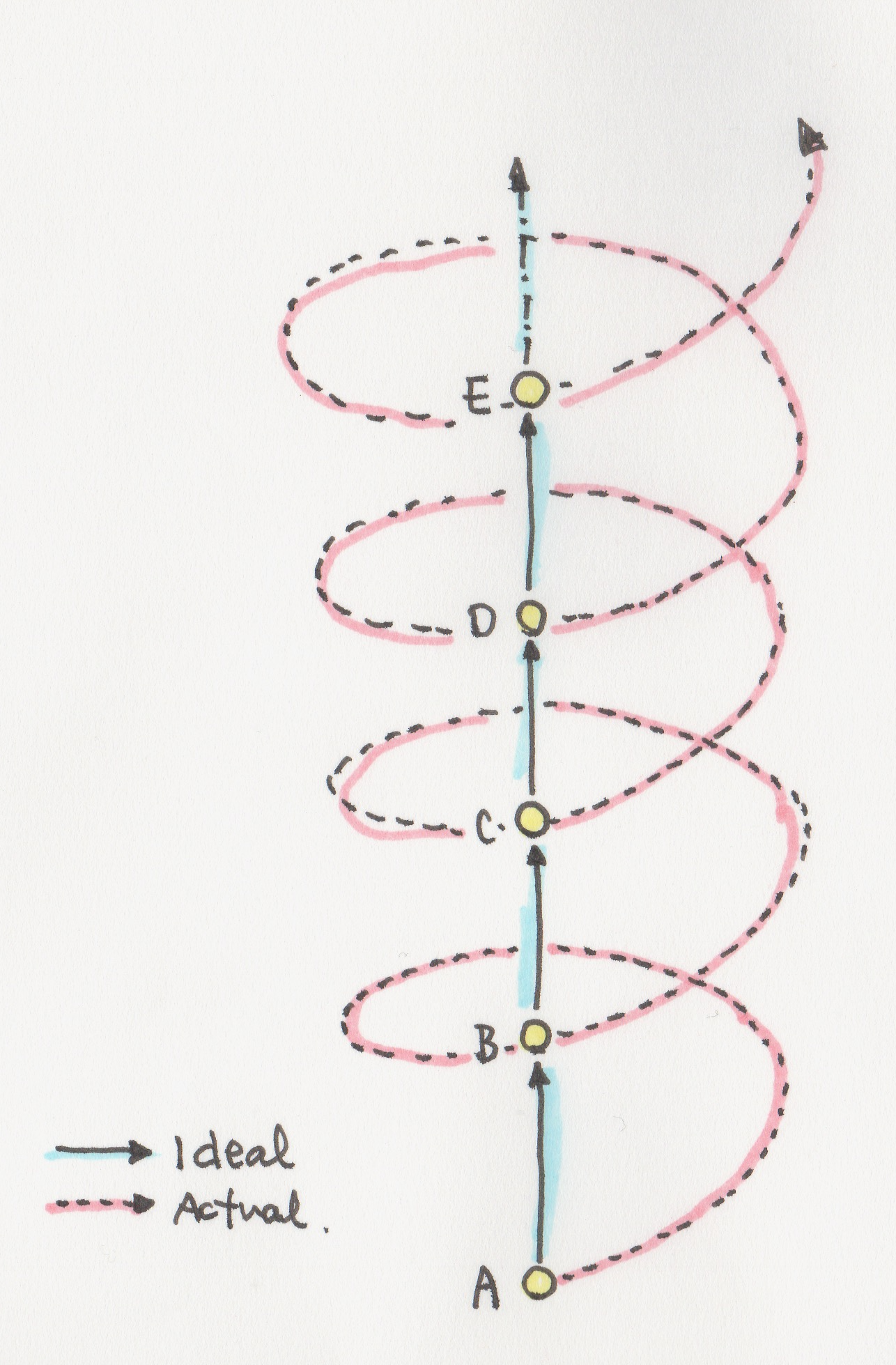
You start somewhere and you go in one direction, and then you realize that you’re not progressing in the right way. And you end up almost back in the same spot, but maybe a bit further. It is slow going directly from point A to point B. It is hard to accept because it feels like you are wasting time.
Oh, that sounds stressful…
Philosophy and mathematics
You seem to have quite a solid way of thinking, why?
I think it’s due to the French education system. We were required to take philosophy classes.
France has had a long tradition of philosophers directly engaged with society’s challenges and evolution, so there’s always been kind of a tight connection between the political reality of France and important philosophical development especially around the 18th century. A further reason for having such a variety of perspectives needing to coexist in France relates to huge variety of people living in it, with different backgrounds, different contexts, different religions, and even different family structures. But they needed to reach a consensus to live together.
Hmm, I see. It’s quite a different history from Japan.
True. So the French people and other European people had to develop new approaches to overcome the challenges emerging from this diversity. One result of this is the variety of traditions found within Europe. For example the strong tradition of rigorous and sophisticated theorization found in Germany, which contrasts with the more discursive and action oriented tradition found in France.
Japanese people overall are not good at either approaches…
One thing that surprised me is that there’s no class for math in the biological programs.
Really? I took it when I was a student, even though it was more than 20 years ago.
Haha. It is true, at least in recent years, as far as I know.
But Japanese people have a strong tradition of spirituality and introspection.
I agree with that. But I now see why Japan is currently struggling to secure enough people for data science and bioinformatics in the life science area.
I think Japan’s educational system needs to generalize theoretical courses, including philosophy and mathematics, so that these reasoning tools can be leveraged by any researcher
Thank you so much. I didn’t expect to talk about many other topics besides science. I enjoyed our talk today.
Thank you.

postscript
I thought he was a bit older than he actually is since he has quite a concrete opinion about whatever we talked about. But as I found out in the interview, he is quite young. It must be because of the differences in the educational system. This interview made me reflect about what we need to do for the upcoming younger generation.
